Cobalt »
PDB 2xwq-3bbi »
3a4j »
Cobalt in PDB 3a4j: Arpte (K185R/D208G/N265D/T274N)
Enzymatic activity of Arpte (K185R/D208G/N265D/T274N)
All present enzymatic activity of Arpte (K185R/D208G/N265D/T274N):
3.1.8.1;
3.1.8.1;
Protein crystallography data
The structure of Arpte (K185R/D208G/N265D/T274N), PDB code: 3a4j
was solved by
J.L.Foo,
C.J.Jackson,
P.D.Carr,
D.L.Ollis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.25 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 108.946, 108.946, 62.884, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.8 / 14.5 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Arpte (K185R/D208G/N265D/T274N)
(pdb code 3a4j). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 2 binding sites of Cobalt where determined in the Arpte (K185R/D208G/N265D/T274N), PDB code: 3a4j:
Jump to Cobalt binding site number: 1; 2;
In total 2 binding sites of Cobalt where determined in the Arpte (K185R/D208G/N265D/T274N), PDB code: 3a4j:
Jump to Cobalt binding site number: 1; 2;
Cobalt binding site 1 out of 2 in 3a4j
Go back to
Cobalt binding site 1 out
of 2 in the Arpte (K185R/D208G/N265D/T274N)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Arpte (K185R/D208G/N265D/T274N) within 5.0Å range:
|
Cobalt binding site 2 out of 2 in 3a4j
Go back to
Cobalt binding site 2 out
of 2 in the Arpte (K185R/D208G/N265D/T274N)
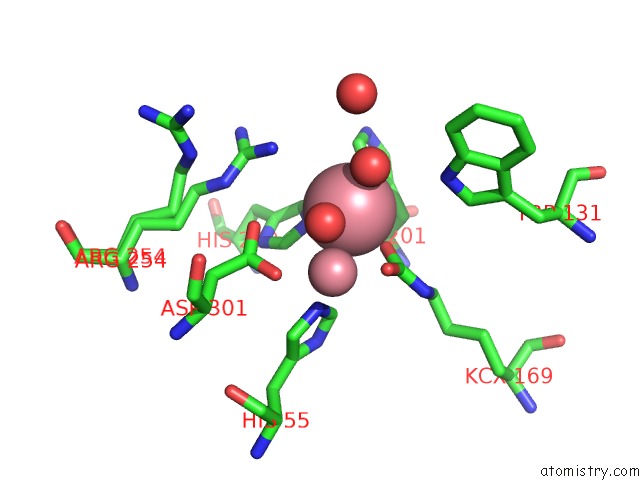
Mono view
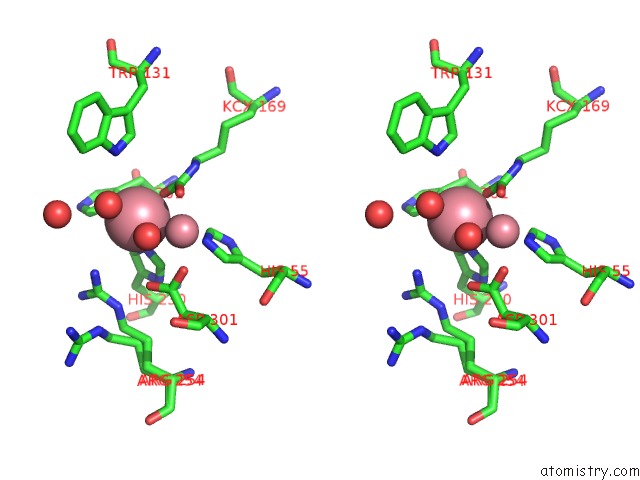
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Arpte (K185R/D208G/N265D/T274N) within 5.0Å range:
|
Reference:
C.J.Jackson,
J.-L.Foo,
N.Tokuriki,
L.Afriat,
P.D.Carr,
H.-K.Kim,
G.Schenk,
D.S.Tawfik,
D.L.Ollis.
Conformational Sampling, Catalysis, and Evolution of the Bacterial Phosphotriesterase Proc.Natl.Acad.Sci.Usa 2009.
ISSN: ESSN 1091-6490
PubMed: 19966226
DOI: 10.1073/PNAS.0907548106
Page generated: Sun Jul 13 18:41:58 2025
ISSN: ESSN 1091-6490
PubMed: 19966226
DOI: 10.1073/PNAS.0907548106
Last articles
Cu in 3T6QCu in 3T5W
Cu in 3T56
Cu in 3T53
Cu in 3T51
Cu in 3T0U
Cu in 3SQR
Cu in 3SOD
Cu in 3SES
Cu in 3SBQ