Cobalt »
PDB 3bbk-3ger »
3fl6 »
Cobalt in PDB 3fl6: Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
Protein crystallography data
The structure of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc), PDB code: 3fl6
was solved by
K.Robeyns,
P.Herdewijn,
L.Van Meervelt,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.86 / 1.17 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 25.720, 33.570, 81.200, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.5 / n/a |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
(pdb code 3fl6). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 8 binding sites of Cobalt where determined in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc), PDB code: 3fl6:
Jump to Cobalt binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Cobalt where determined in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc), PDB code: 3fl6:
Jump to Cobalt binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Cobalt binding site 1 out of 8 in 3fl6
Go back to
Cobalt binding site 1 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
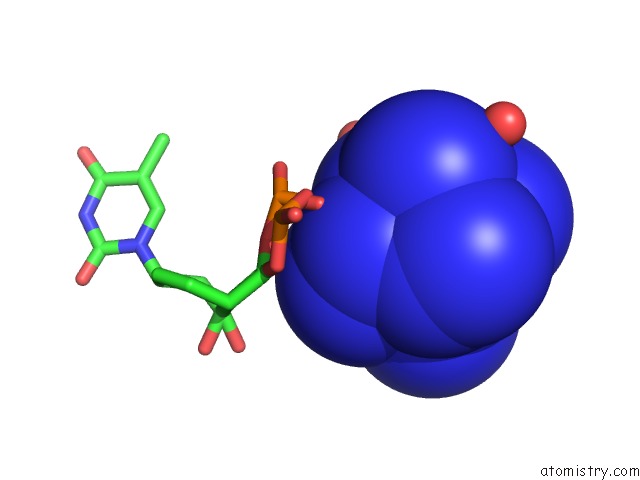
Mono view
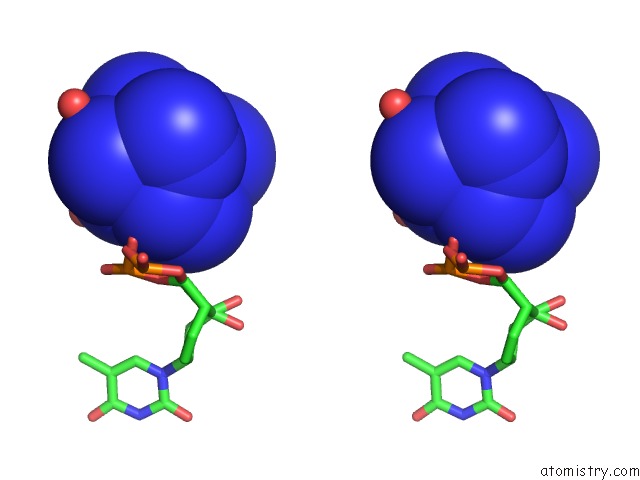
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 2 out of 8 in 3fl6
Go back to
Cobalt binding site 2 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
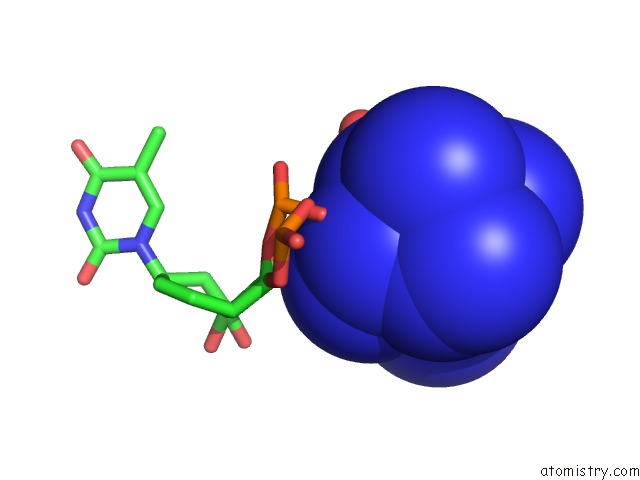
Mono view
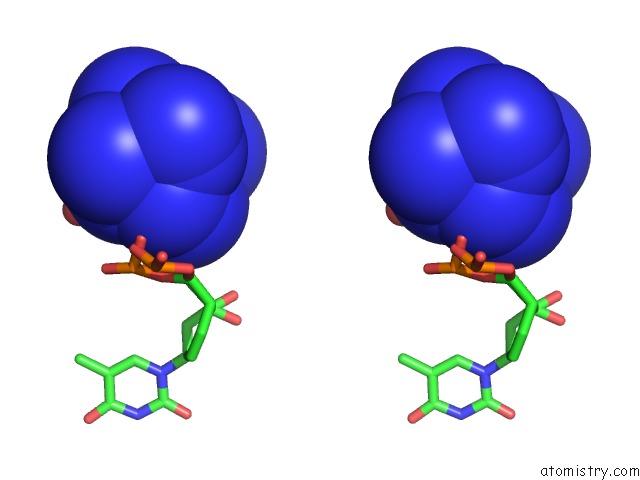
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 3 out of 8 in 3fl6
Go back to
Cobalt binding site 3 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
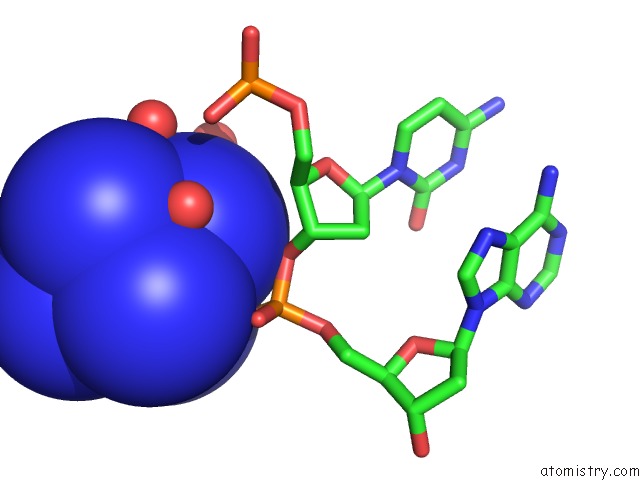
Mono view
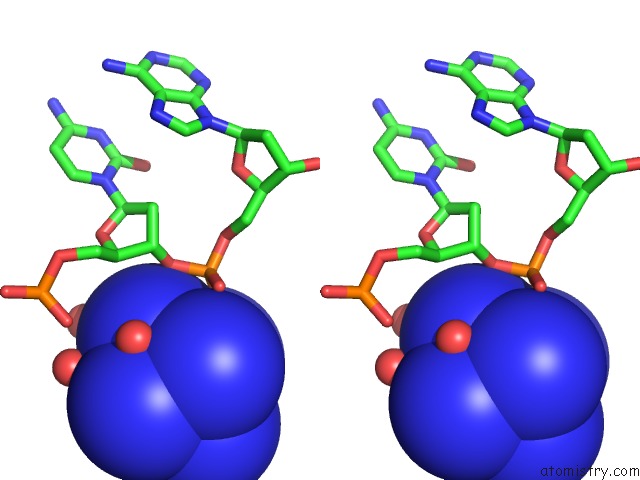
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 3 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 4 out of 8 in 3fl6
Go back to
Cobalt binding site 4 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
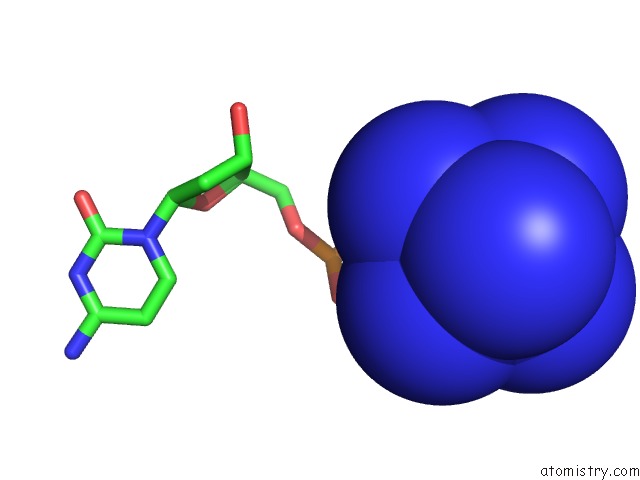
Mono view
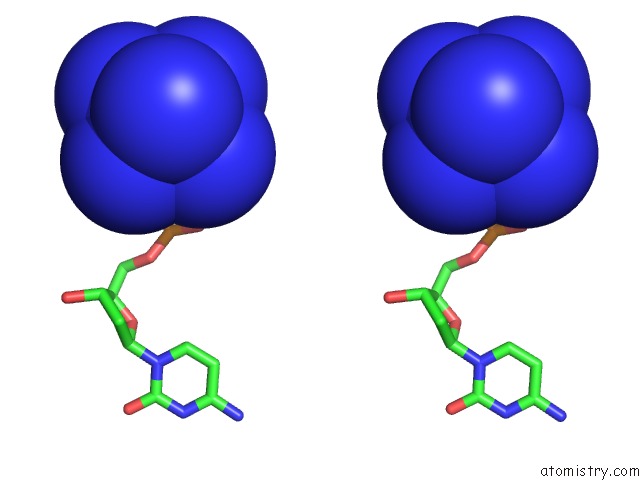
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 4 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 5 out of 8 in 3fl6
Go back to
Cobalt binding site 5 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
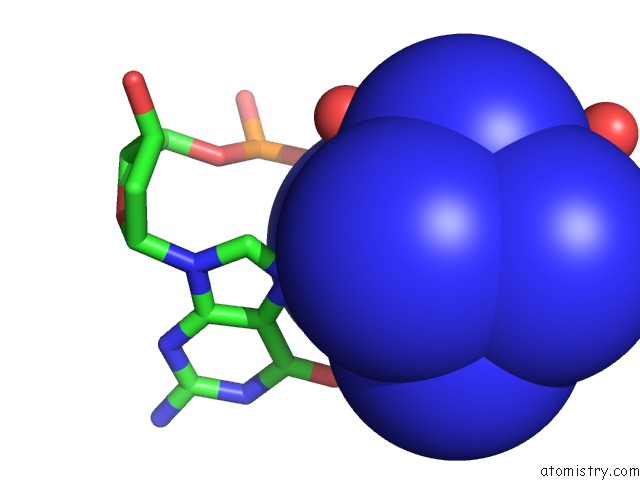
Mono view
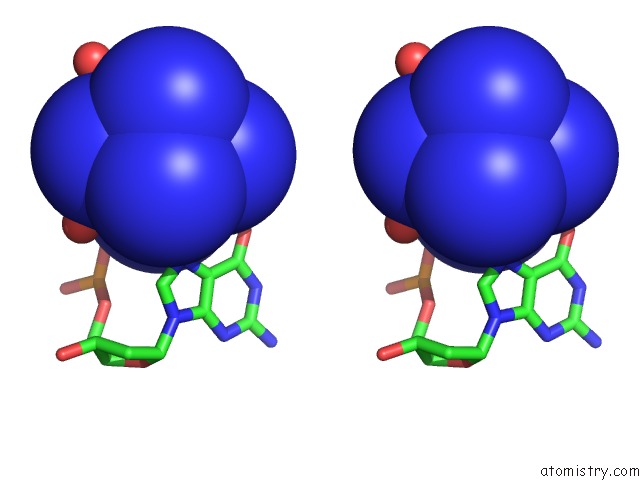
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 5 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 6 out of 8 in 3fl6
Go back to
Cobalt binding site 6 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 6 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 7 out of 8 in 3fl6
Go back to
Cobalt binding site 7 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
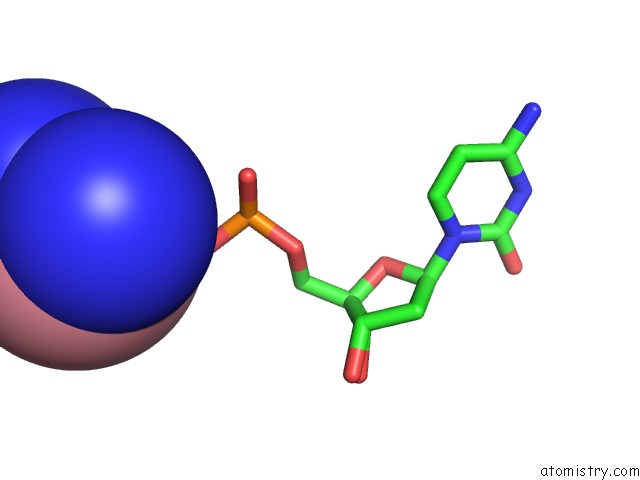
Mono view
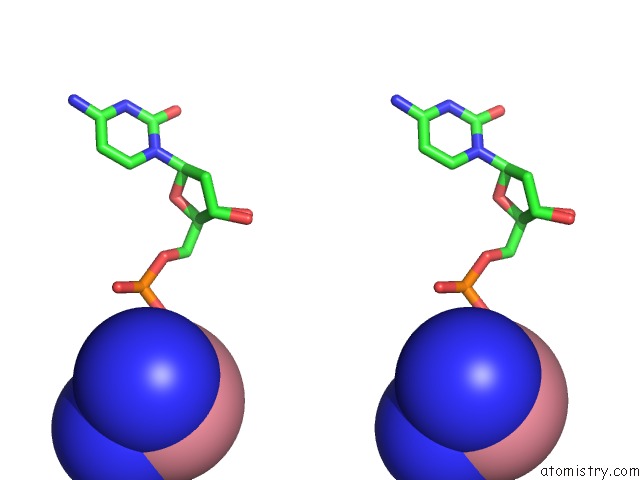
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 7 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Cobalt binding site 8 out of 8 in 3fl6
Go back to
Cobalt binding site 8 out
of 8 in the Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc)
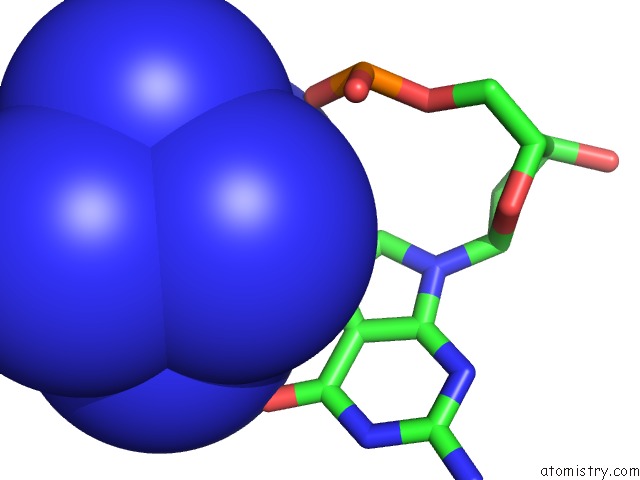
Mono view
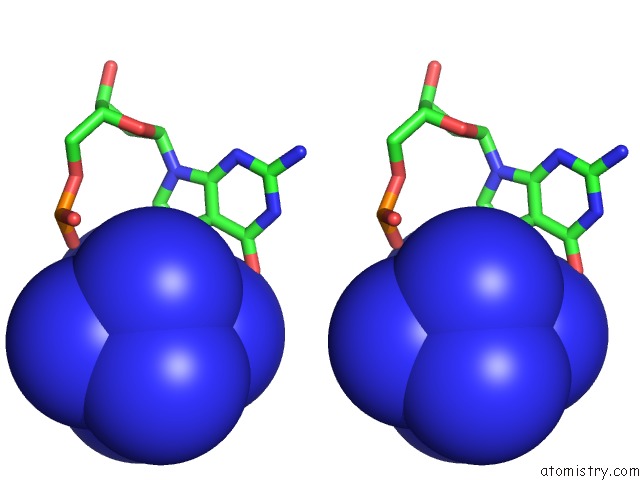
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 8 of Influence of the Incorporation of A Cyclohexenyl Nucleic Acid (Cena) Residue Onto the Sequence D(Gcgtgcg)/D(Cgcacgc) within 5.0Å range:
|
Reference:
K.Robeyns,
P.Herdewijn,
L.Van Meervelt.
Direct Observation of Two Cyclohexenyl (Cena) Ring Conformations in Duplex Dna. Artif Dna Pna Xna V. 1 2 2010.
ISSN: ESSN 1949-0968
PubMed: 21687521
DOI: 10.4161/ADNA.1.1.10952
Page generated: Sun Jul 13 18:50:32 2025
ISSN: ESSN 1949-0968
PubMed: 21687521
DOI: 10.4161/ADNA.1.1.10952
Last articles
Cu in 3S3BCu in 3S3A
Cu in 3S39
Cu in 3S38
Cu in 3RGB
Cu in 3S0P
Cu in 3S33
Cu in 3RFR
Cu in 3QVZ
Cu in 3QPK