Cobalt »
PDB 5d6f-5ikv »
5eje »
Cobalt in PDB 5eje: Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A
Enzymatic activity of Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A
All present enzymatic activity of Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A:
2.7.4.3;
2.7.4.3;
Protein crystallography data
The structure of Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A, PDB code: 5eje
was solved by
U.H.Sauer,
M.Kovermann,
C.Grundstrom,
M.Wolf-Watz,
A.E.Sauer-Eriksson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.80 / 1.90 |
| Space group | P 21 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.003, 79.064, 81.834, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.6 / 23.6 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A
(pdb code 5eje). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 2 binding sites of Cobalt where determined in the Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A, PDB code: 5eje:
Jump to Cobalt binding site number: 1; 2;
In total 2 binding sites of Cobalt where determined in the Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A, PDB code: 5eje:
Jump to Cobalt binding site number: 1; 2;
Cobalt binding site 1 out of 2 in 5eje
Go back to
Cobalt binding site 1 out
of 2 in the Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A
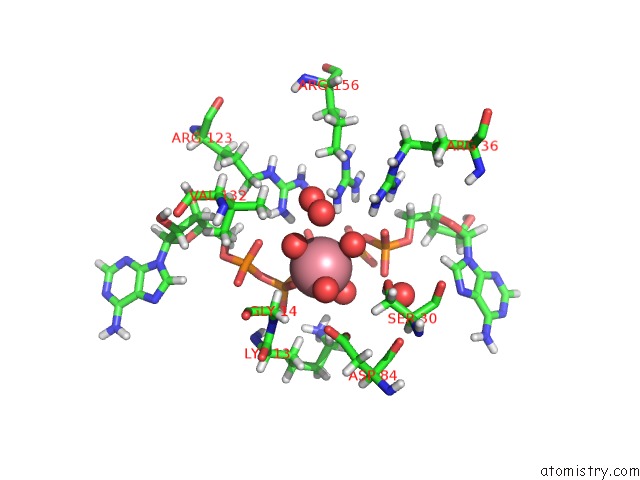
Mono view
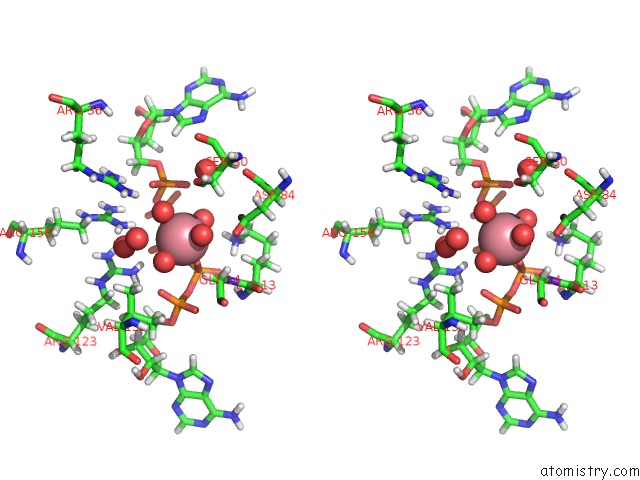
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A within 5.0Å range:
|
Cobalt binding site 2 out of 2 in 5eje
Go back to
Cobalt binding site 2 out
of 2 in the Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A
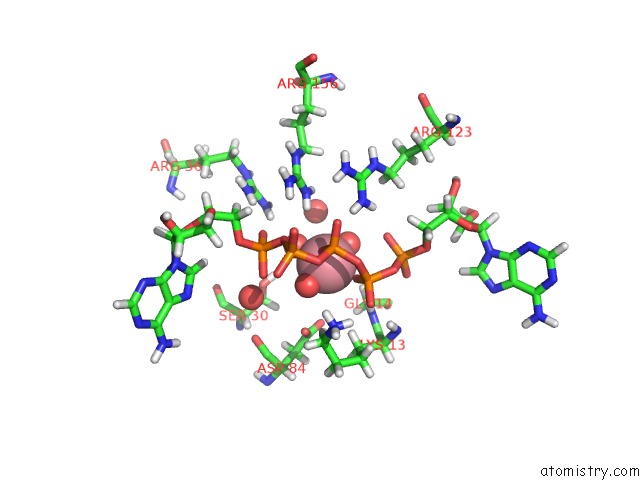
Mono view
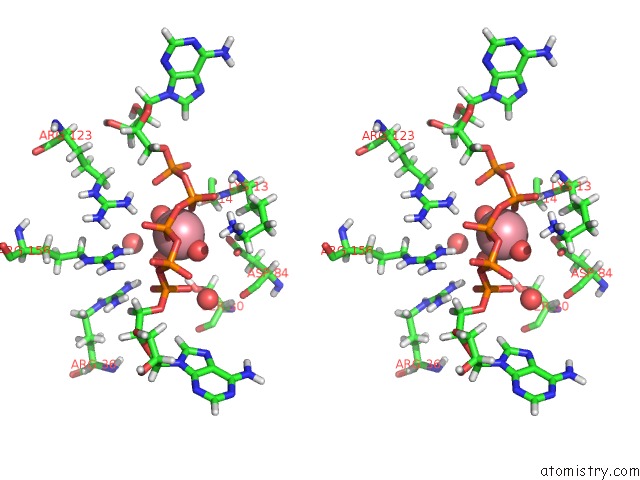
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Crystal Structure of E. Coli Adenylate Kinase G56C/T163C Double Mutant in Complex with AP5A within 5.0Å range:
|
Reference:
M.Kovermann,
C.Grundstrom,
A.E.Sauer-Eriksson,
U.H.Sauer,
M.Wolf-Watz.
Structural Basis For Ligand Binding to An Enzyme By A Conformational Selection Pathway. Proc. Natl. Acad. Sci. V. 114 6298 2017U.S.A..
ISSN: ESSN 1091-6490
PubMed: 28559350
DOI: 10.1073/PNAS.1700919114
Page generated: Sun Jul 13 20:20:59 2025
ISSN: ESSN 1091-6490
PubMed: 28559350
DOI: 10.1073/PNAS.1700919114
Last articles
Cu in 6UWECu in 6VBS
Cu in 6VBT
Cu in 6TTD
Cu in 6TYR
Cu in 6UTR
Cu in 6U2Z
Cu in 6TWE
Cu in 6TFO
Cu in 6TFD