Cobalt »
PDB 5d6f-5ikv »
5iav »
Cobalt in PDB 5iav: Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
Protein crystallography data
The structure of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav
was solved by
N.J.Schnicker,
M.Dey,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.68 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.769, 41.495, 111.043, 90.00, 96.67, 90.00 |
| R / Rfree (%) | 21.2 / 22.9 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
(pdb code 5iav). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 2 binding sites of Cobalt where determined in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav:
Jump to Cobalt binding site number: 1; 2;
In total 2 binding sites of Cobalt where determined in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav:
Jump to Cobalt binding site number: 1; 2;
Cobalt binding site 1 out of 2 in 5iav
Go back to
Cobalt binding site 1 out
of 2 in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
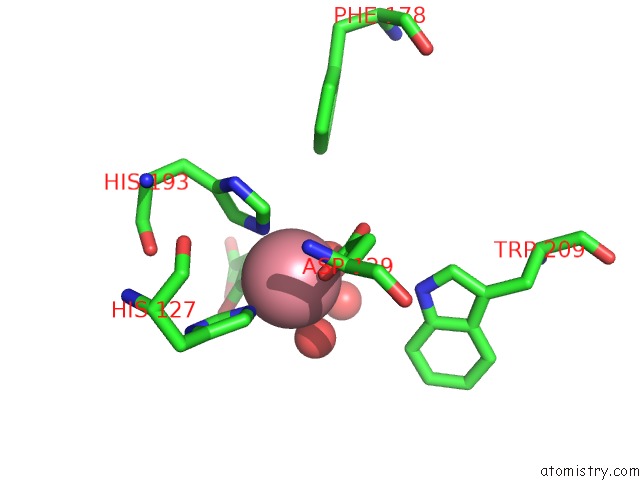
Mono view
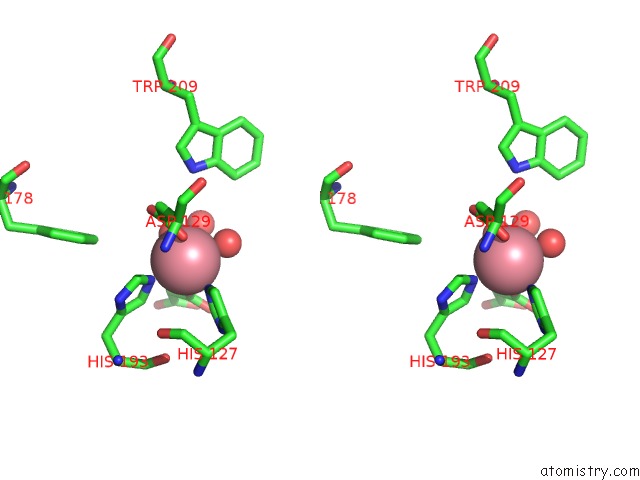
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli within 5.0Å range:
|
Cobalt binding site 2 out of 2 in 5iav
Go back to
Cobalt binding site 2 out
of 2 in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
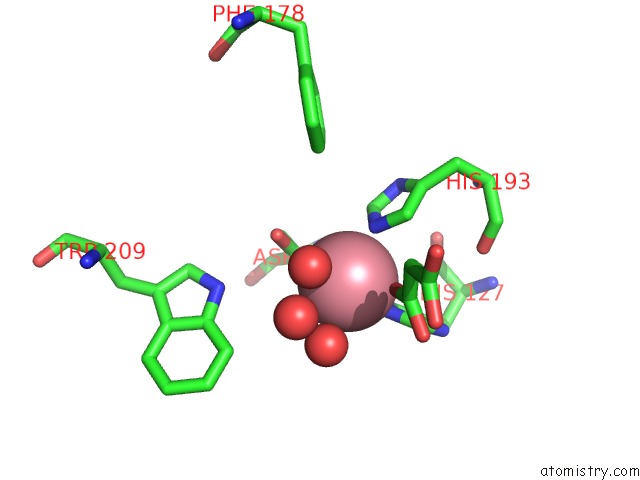
Mono view
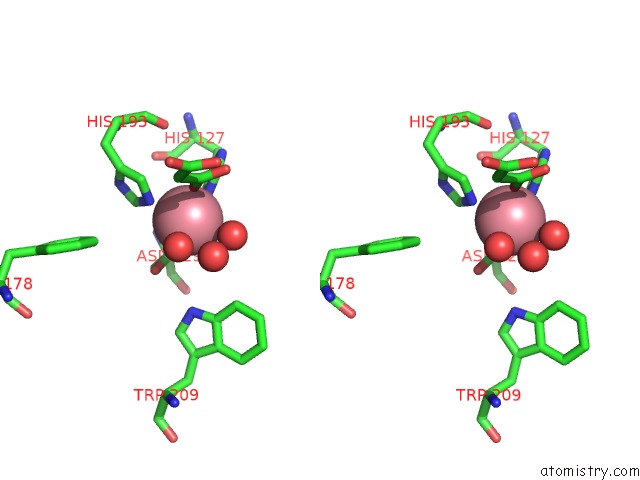
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli within 5.0Å range:
|
Reference:
N.J.Schnicker,
M.Dey.
Bacillus Anthracis Prolyl 4-Hydroxylase Modifies Collagen-Like Substrates in Asymmetric Patterns. J.Biol.Chem. V. 291 13360 2016.
ISSN: ESSN 1083-351X
PubMed: 27129244
DOI: 10.1074/JBC.M116.725432
Page generated: Sun Jul 13 20:26:20 2025
ISSN: ESSN 1083-351X
PubMed: 27129244
DOI: 10.1074/JBC.M116.725432
Last articles
Cu in 6RGPCu in 6RGH
Cu in 6R01
Cu in 6REK
Cu in 6REH
Cu in 6R1L
Cu in 6QVH
Cu in 6QXD
Cu in 6QWW
Cu in 6QUG