Cobalt »
PDB 6kgh-6oxc »
6m2g »
Cobalt in PDB 6m2g: Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin
Enzymatic activity of Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin
All present enzymatic activity of Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin:
4.99.1.11; 4.99.1.3;
4.99.1.11; 4.99.1.3;
Protein crystallography data
The structure of Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin, PDB code: 6m2g
was solved by
T.Fujishiro,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.92 / 2.80 |
| Space group | P 41 |
| Cell size a, b, c (Å), α, β, γ (°) | 69.18, 69.18, 81.8, 90, 90, 90 |
| R / Rfree (%) | 18.4 / 22.4 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin
(pdb code 6m2g). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin, PDB code: 6m2g:
In total only one binding site of Cobalt was determined in the Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin, PDB code: 6m2g:
Cobalt binding site 1 out of 1 in 6m2g
Go back to
Cobalt binding site 1 out
of 1 in the Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin
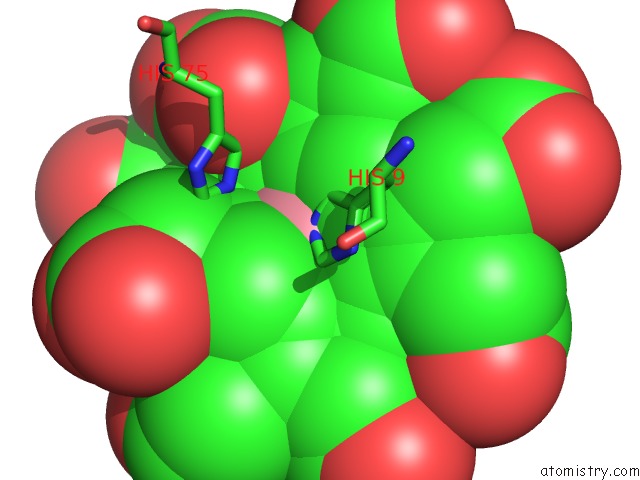
Mono view
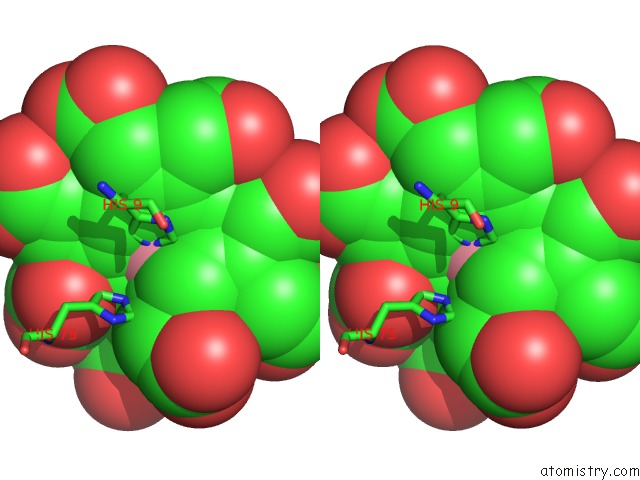
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Sirohydrochlorin Nickelochelatase Cfba in Complex with Cobalt- Sirohydrochlorin within 5.0Å range:
|
Reference:
T.Fujishiro,
S.Ogawa.
The Nickel-Sirohydrochlorin Formation Mechanismof the Ancestral Class II Chelatase Cfba in COENZYMEF430 Biosynthesis Chem Sci 2021.
ISSN: ESSN 2041-6539
DOI: 10.1039/D0SC05439A
Page generated: Tue Jul 30 18:51:34 2024
ISSN: ESSN 2041-6539
DOI: 10.1039/D0SC05439A
Last articles
As in 4O55As in 4OSG
As in 4O0J
As in 4NLY
As in 4NLX
As in 4NSE
As in 4NLU
As in 4NLW
As in 4NLV
As in 4NLT