Cobalt »
PDB 8im2-8sfb »
8jbw »
Cobalt in PDB 8jbw: Crystal Structure of Zthppd-(+)-Usnic Acid Complex
Enzymatic activity of Crystal Structure of Zthppd-(+)-Usnic Acid Complex
All present enzymatic activity of Crystal Structure of Zthppd-(+)-Usnic Acid Complex:
1.13.11.27;
1.13.11.27;
Protein crystallography data
The structure of Crystal Structure of Zthppd-(+)-Usnic Acid Complex, PDB code: 8jbw
was solved by
H.-Y.Lin,
J.Dong,
G.-F.Yang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.35 / 2.65 |
| Space group | P 42 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 110.348, 110.348, 80.849, 90, 90, 90 |
| R / Rfree (%) | 25.3 / 30.3 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Crystal Structure of Zthppd-(+)-Usnic Acid Complex
(pdb code 8jbw). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Crystal Structure of Zthppd-(+)-Usnic Acid Complex, PDB code: 8jbw:
In total only one binding site of Cobalt was determined in the Crystal Structure of Zthppd-(+)-Usnic Acid Complex, PDB code: 8jbw:
Cobalt binding site 1 out of 1 in 8jbw
Go back to
Cobalt binding site 1 out
of 1 in the Crystal Structure of Zthppd-(+)-Usnic Acid Complex
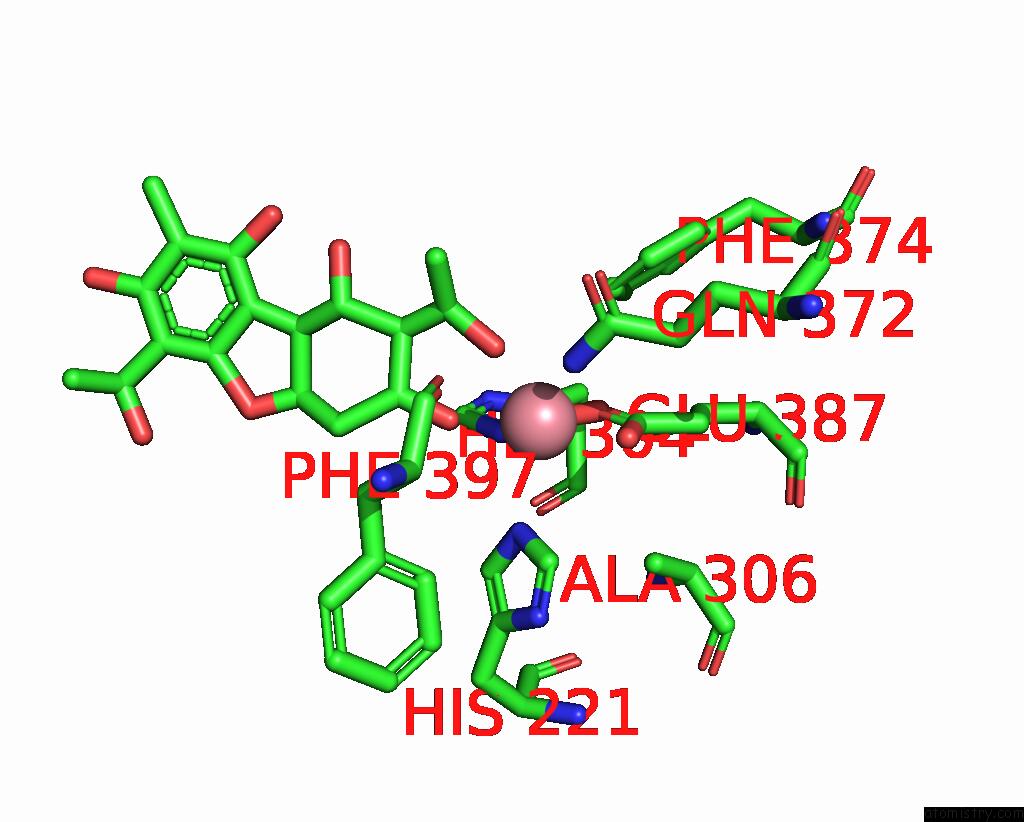
Mono view
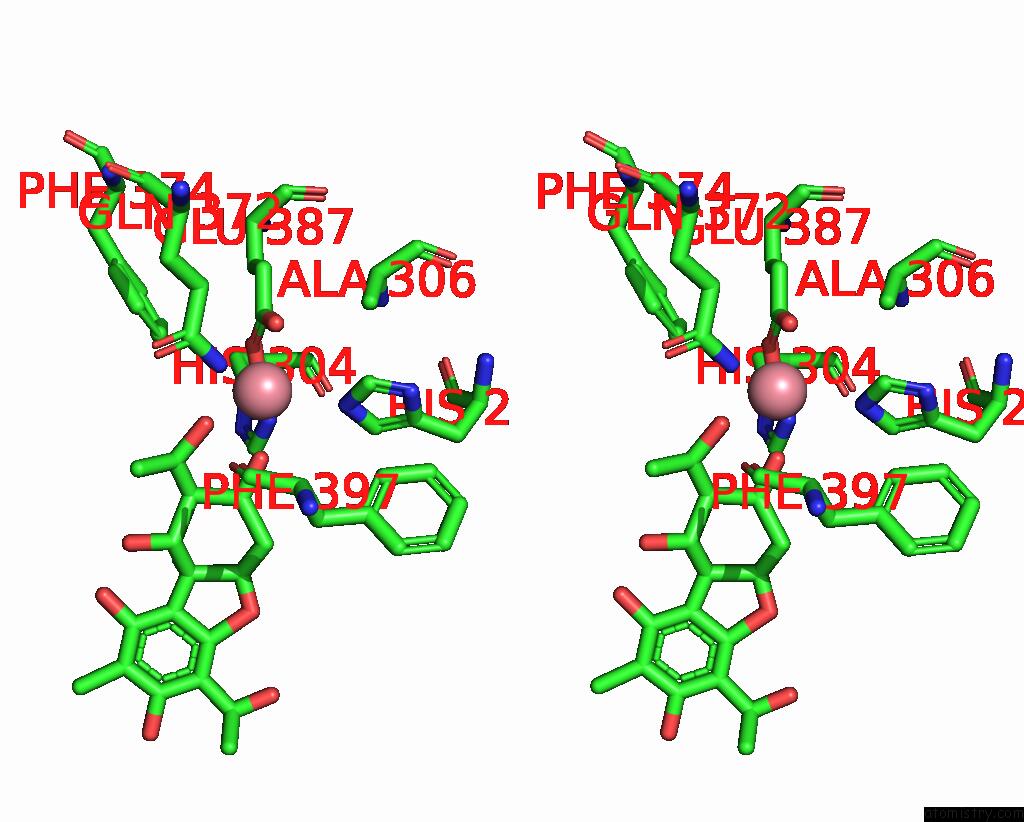
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Crystal Structure of Zthppd-(+)-Usnic Acid Complex within 5.0Å range:
|
Reference:
X.H.Yu,
J.Dong,
C.P.Fan,
M.X.Chen,
M.Li,
B.F.Zheng,
Y.F.Hu,
H.Y.Lin,
G.F.Yang.
Discovery and Development of 4-Hydroxyphenylpyruvate Dioxygenase As A Novel Crop Fungicide Target. J.Agric.Food Chem. V. 71 19396 2023.
ISSN: ESSN 1520-5118
PubMed: 38035573
DOI: 10.1021/ACS.JAFC.3C05260
Page generated: Sun Jul 13 22:01:18 2025
ISSN: ESSN 1520-5118
PubMed: 38035573
DOI: 10.1021/ACS.JAFC.3C05260
Last articles
Cu in 5ZHWCu in 5ZCP
Cu in 5ZCQ
Cu in 5Z86
Cu in 5ZCO
Cu in 5Z85
Cu in 5Z84
Cu in 5Z1X
Cu in 5Z22
Cu in 5Z62