Cobalt »
PDB 8im2-8sfb »
8sa4 »
Cobalt in PDB 8sa4: Adenosylcobalamin-Bound Riboswitch Dimer, Form 3
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Adenosylcobalamin-Bound Riboswitch Dimer, Form 3
(pdb code 8sa4). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Adenosylcobalamin-Bound Riboswitch Dimer, Form 3, PDB code: 8sa4:
In total only one binding site of Cobalt was determined in the Adenosylcobalamin-Bound Riboswitch Dimer, Form 3, PDB code: 8sa4:
Cobalt binding site 1 out of 1 in 8sa4
Go back to
Cobalt binding site 1 out
of 1 in the Adenosylcobalamin-Bound Riboswitch Dimer, Form 3
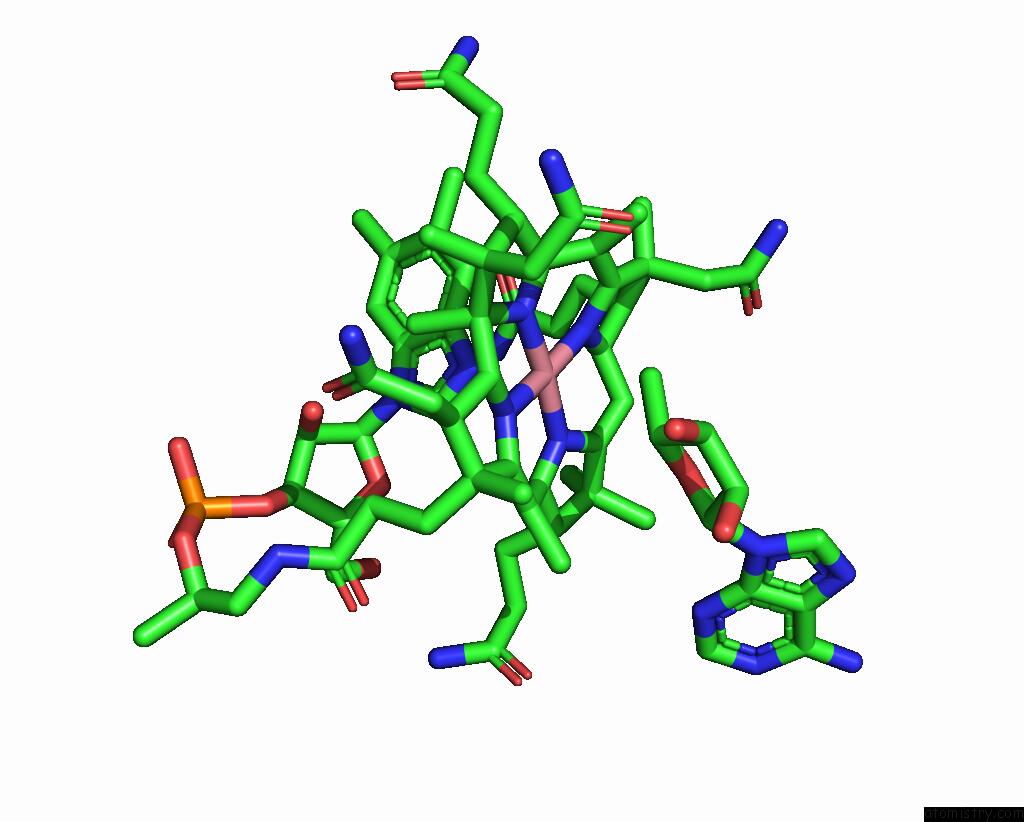
Mono view
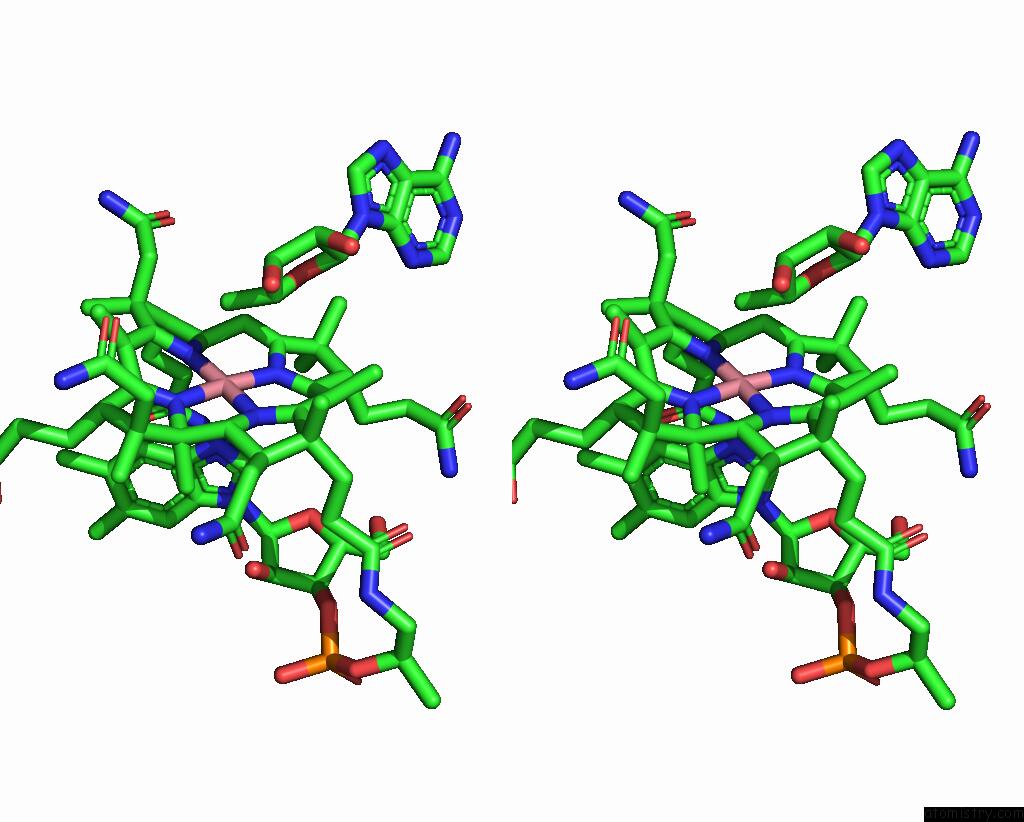
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Adenosylcobalamin-Bound Riboswitch Dimer, Form 3 within 5.0Å range:
|
Reference:
J.Ding,
J.C.Deme,
J.R.Stagno,
P.Yu,
S.M.Lea,
Y.X.Wang.
Capturing Heterogeneous Conformers of Cobalamin Riboswitch By Cryo-Em Nucleic Acids Res. 2023.
ISSN: ESSN 1362-4962
Page generated: Sun Jul 13 22:03:59 2025
ISSN: ESSN 1362-4962
Last articles
Cu in 6REHCu in 6R1L
Cu in 6QVH
Cu in 6QXD
Cu in 6QWW
Cu in 6QUG
Cu in 6QWG
Cu in 6QUH
Cu in 6QUJ
Cu in 6QUI