Cobalt »
PDB 1nyi-1rr2 »
1ofa »
Cobalt in PDB 1ofa: Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II)
Enzymatic activity of Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II)
All present enzymatic activity of Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II):
4.1.2.15;
4.1.2.15;
Protein crystallography data
The structure of Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II), PDB code: 1ofa
was solved by
V.Koenig,
A.Pfeil,
G.Heinrich,
G.H.Braus,
T.R.Schneider,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 17.57 / 2.01 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 196.210, 50.610, 64.980, 90.00, 106.33, 90.00 |
| R / Rfree (%) | 15.5 / 20.4 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II)
(pdb code 1ofa). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 2 binding sites of Cobalt where determined in the Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II), PDB code: 1ofa:
Jump to Cobalt binding site number: 1; 2;
In total 2 binding sites of Cobalt where determined in the Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II), PDB code: 1ofa:
Jump to Cobalt binding site number: 1; 2;
Cobalt binding site 1 out of 2 in 1ofa
Go back to
Cobalt binding site 1 out
of 2 in the Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II)
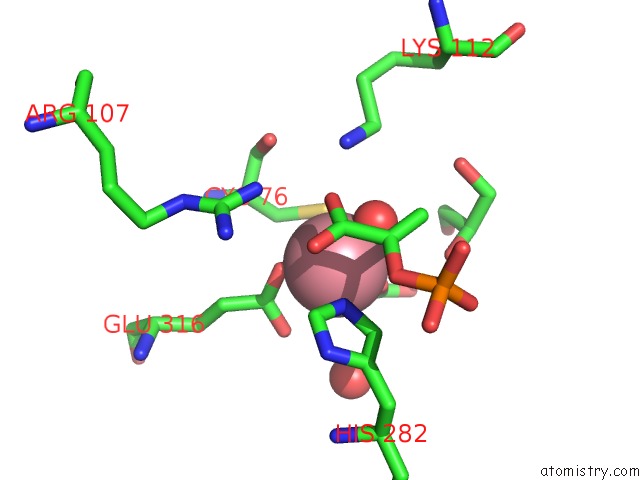
Mono view
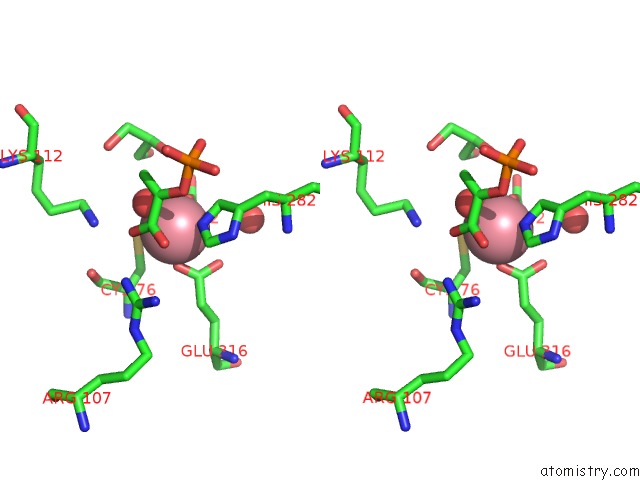
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II) within 5.0Å range:
|
Cobalt binding site 2 out of 2 in 1ofa
Go back to
Cobalt binding site 2 out
of 2 in the Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II)
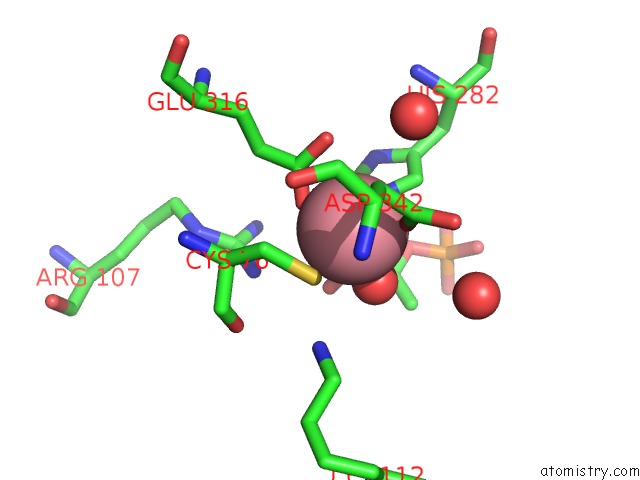
Mono view
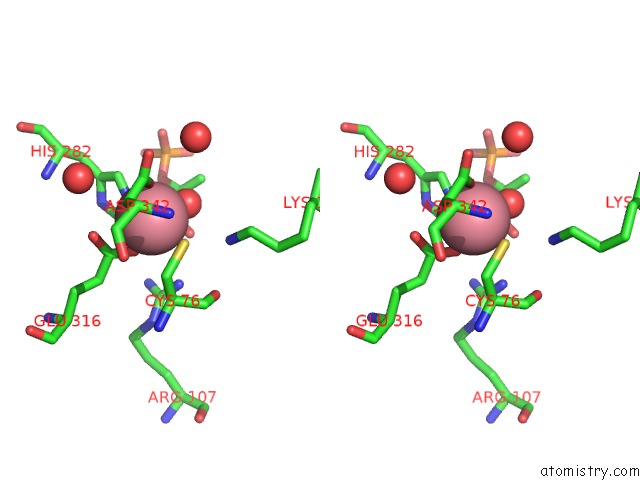
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Crystal Structure of the Tyrosine-Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae in Complex with Phosphoenolpyruvate and Cobalt(II) within 5.0Å range:
|
Reference:
V.Koenig,
A.Pfeil,
G.H.Braus,
T.R.Schneider.
Substrate and Metal Complexes of 3-Deoxy-D- Arabino-Heptulosonate-7-Phosphate Synthase From Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism J.Mol.Biol. V. 337 675 2004.
ISSN: ISSN 0022-2836
PubMed: 15019786
DOI: 10.1016/J.JMB.2004.01.055
Page generated: Sun Jul 13 17:40:48 2025
ISSN: ISSN 0022-2836
PubMed: 15019786
DOI: 10.1016/J.JMB.2004.01.055
Last articles
K in 8HIRK in 8GYR
K in 8GR9
K in 8H5L
K in 8H5J
K in 8GPE
K in 8GNG
K in 8GPD
K in 8GJB
K in 8G7N