Cobalt »
PDB 5d6f-5ikv »
5iav »
Cobalt in PDB 5iav: Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
Protein crystallography data
The structure of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav
was solved by
N.J.Schnicker,
M.Dey,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.68 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 42.769, 41.495, 111.043, 90.00, 96.67, 90.00 |
| R / Rfree (%) | 21.2 / 22.9 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
(pdb code 5iav). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 2 binding sites of Cobalt where determined in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav:
Jump to Cobalt binding site number: 1; 2;
In total 2 binding sites of Cobalt where determined in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli, PDB code: 5iav:
Jump to Cobalt binding site number: 1; 2;
Cobalt binding site 1 out of 2 in 5iav
Go back to
Cobalt binding site 1 out
of 2 in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
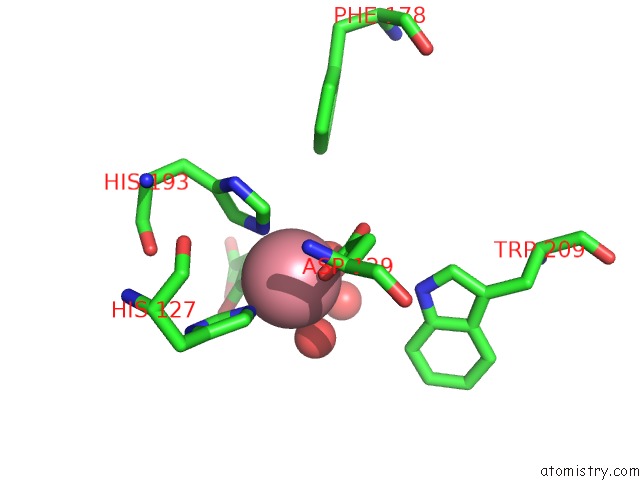
Mono view
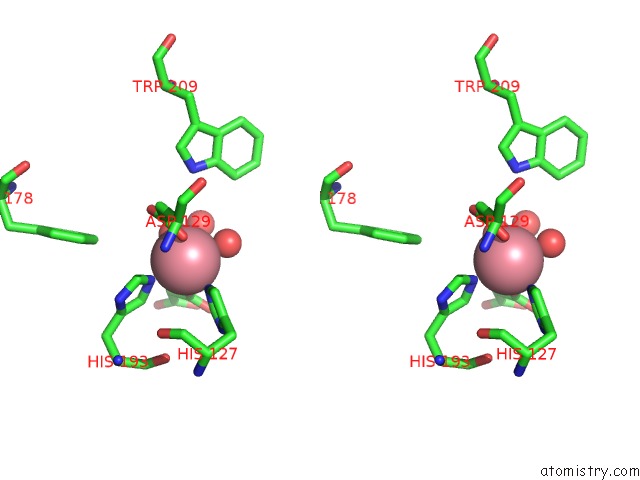
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli within 5.0Å range:
|
Cobalt binding site 2 out of 2 in 5iav
Go back to
Cobalt binding site 2 out
of 2 in the Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli
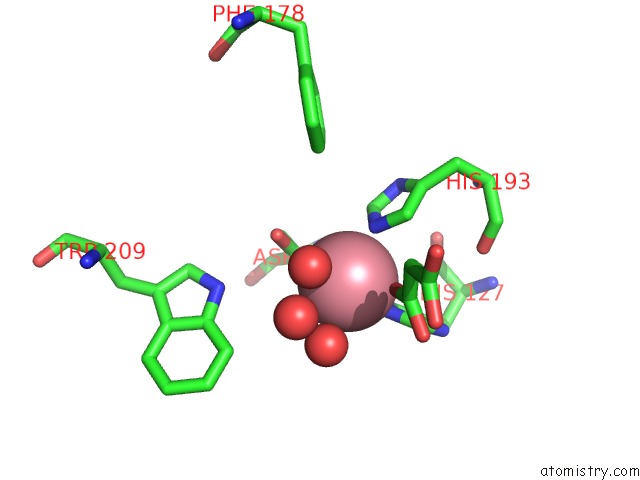
Mono view
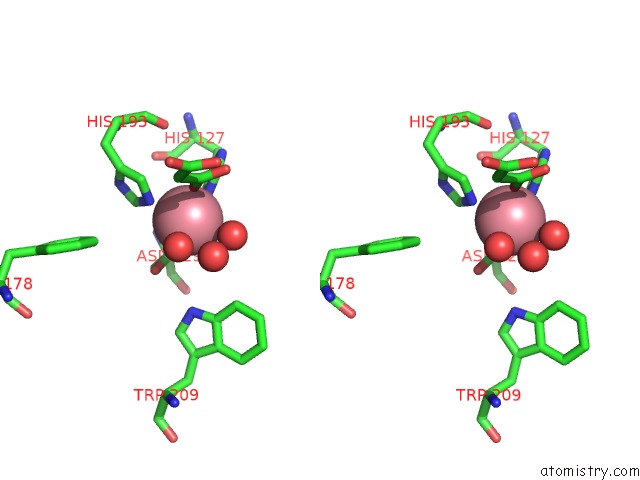
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Mechanistic and Structural Analysis of Substrate Recognition and Cofactor Binding By An Unusual Bacterial Prolyl Hydroxylase - Co- BAP4H-Mli within 5.0Å range:
|
Reference:
N.J.Schnicker,
M.Dey.
Bacillus Anthracis Prolyl 4-Hydroxylase Modifies Collagen-Like Substrates in Asymmetric Patterns. J.Biol.Chem. V. 291 13360 2016.
ISSN: ESSN 1083-351X
PubMed: 27129244
DOI: 10.1074/JBC.M116.725432
Page generated: Sun Jul 13 20:26:20 2025
ISSN: ESSN 1083-351X
PubMed: 27129244
DOI: 10.1074/JBC.M116.725432
Last articles
K in 4R33K in 4R2C
K in 4QXG
K in 4QRH
K in 4QNE
K in 4QGC
K in 4QKA
K in 4QE9
K in 4QG8
K in 4QK8