Cobalt »
PDB 6kgh-6oxc »
6kgw »
Cobalt in PDB 6kgw: Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin
Protein crystallography data
The structure of Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin, PDB code: 6kgw
was solved by
Z.K.Lu,
A.L.Zhang,
X.Liu,
L.Guddat,
H.T.Yang,
Z.H.Rao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.80 / 2.41 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.246, 84.219, 90.527, 90.00, 111.31, 90.00 |
| R / Rfree (%) | 19.7 / 25 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin
(pdb code 6kgw). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 3 binding sites of Cobalt where determined in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin, PDB code: 6kgw:
Jump to Cobalt binding site number: 1; 2; 3;
In total 3 binding sites of Cobalt where determined in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin, PDB code: 6kgw:
Jump to Cobalt binding site number: 1; 2; 3;
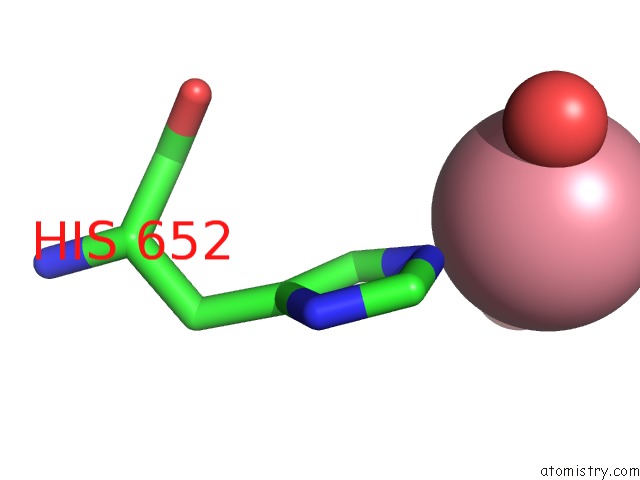
Cobalt binding site 1 out of 3 in 6kgw
Go back to
Cobalt binding site 1 out
of 3 in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin within 5.0Å range:
|
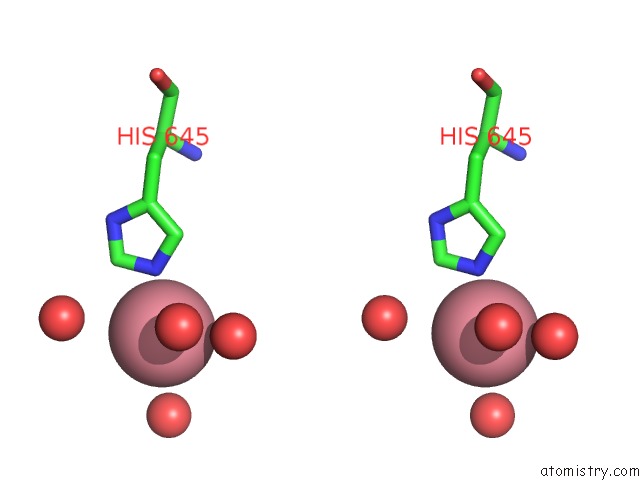
Cobalt binding site 2 out of 3 in 6kgw
Go back to
Cobalt binding site 2 out
of 3 in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin within 5.0Å range:
|
Cobalt binding site 3 out of 3 in 6kgw
Go back to
Cobalt binding site 3 out
of 3 in the Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 3 of Crystal Structure of Penicillin Binding Protein 3 (PBP3) From Mycobacterium Tuerculosis, Complexed with Ampicillin within 5.0Å range:
|
Reference:
Z.Lu,
H.Wang,
A.Zhang,
X.Liu,
W.Zhou,
C.Yang,
L.Guddat,
H.Yang,
C.J.Schofield,
Z.Rao.
Structures Ofmycobacterium Tuberculosispenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation. Mol.Pharmacol. V. 97 287 2020.
ISSN: ESSN 1521-0111
PubMed: 32086254
DOI: 10.1124/MOL.119.118042
Page generated: Sun Jul 13 21:06:49 2025
ISSN: ESSN 1521-0111
PubMed: 32086254
DOI: 10.1124/MOL.119.118042
Last articles
I in 8D22I in 8CXW
I in 8D00
I in 8CT8
I in 8CUF
I in 8CJ7
I in 8CNZ
I in 8C3P
I in 8CEV
I in 8C7R