Cobalt »
PDB 8sfc-8xc3 »
8tcr »
Cobalt in PDB 8tcr: Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
Protein crystallography data
The structure of Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1, PDB code: 8tcr
was solved by
A.C.Lazarski,
L.J.Worrall,
N.C.J.Strynadka,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.40 / 2.08 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 47.517, 113.679, 208.58, 90, 90, 90 |
| R / Rfree (%) | 18.2 / 22.9 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
(pdb code 8tcr). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total 4 binding sites of Cobalt where determined in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1, PDB code: 8tcr:
Jump to Cobalt binding site number: 1; 2; 3; 4;
In total 4 binding sites of Cobalt where determined in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1, PDB code: 8tcr:
Jump to Cobalt binding site number: 1; 2; 3; 4;
Cobalt binding site 1 out of 4 in 8tcr
Go back to
Cobalt binding site 1 out
of 4 in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
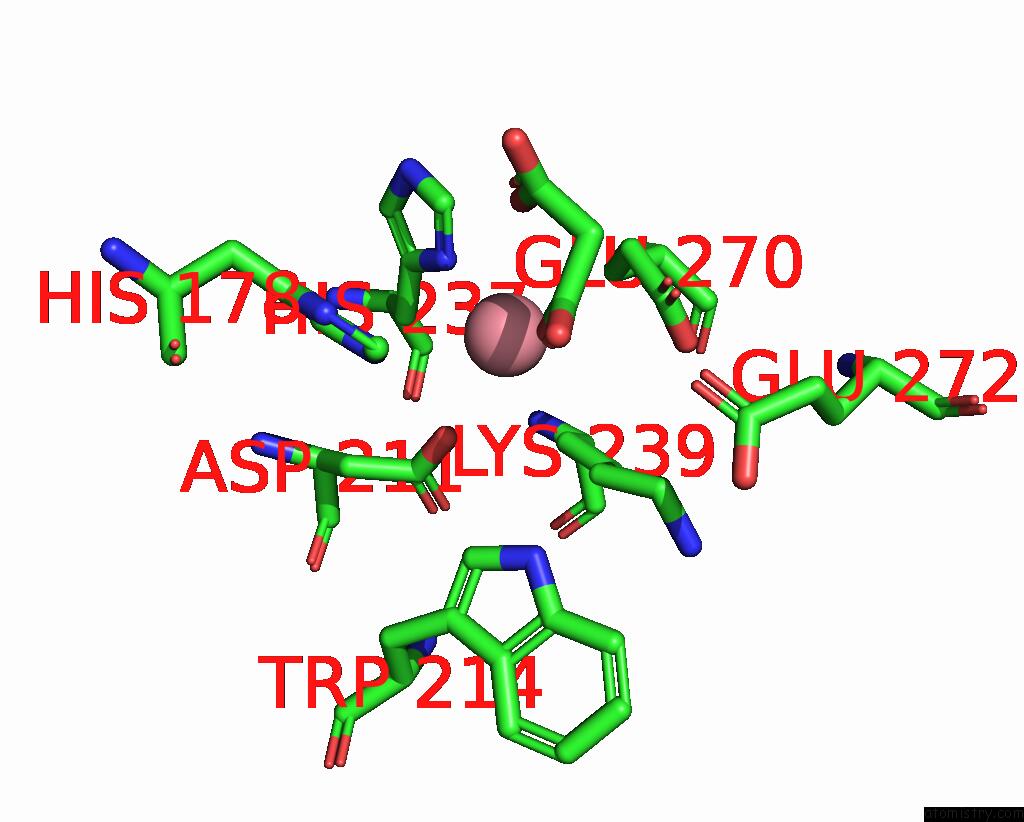
Mono view
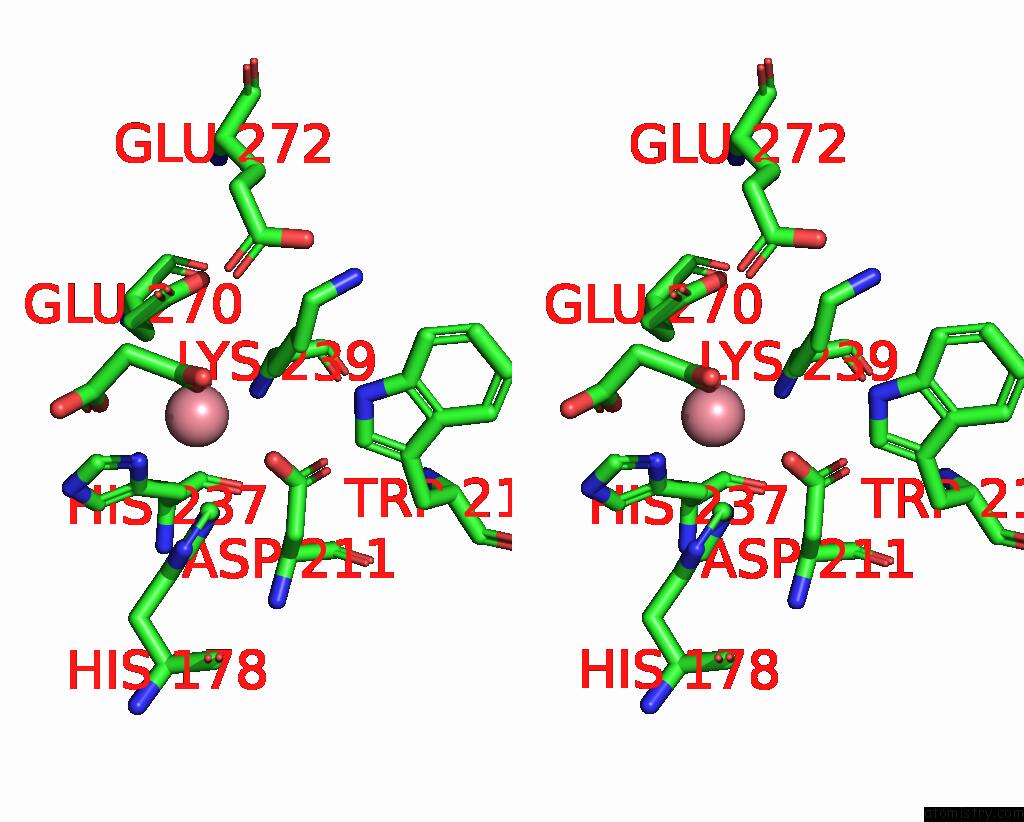
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1 within 5.0Å range:
|
Cobalt binding site 2 out of 4 in 8tcr
Go back to
Cobalt binding site 2 out
of 4 in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
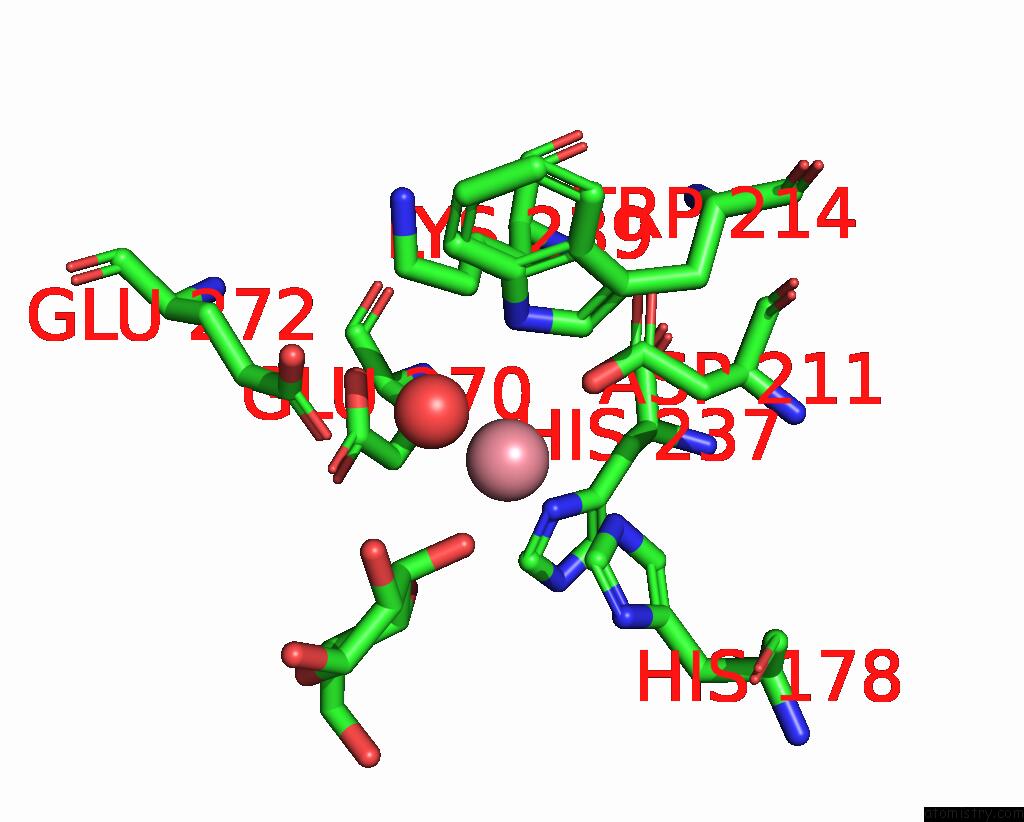
Mono view
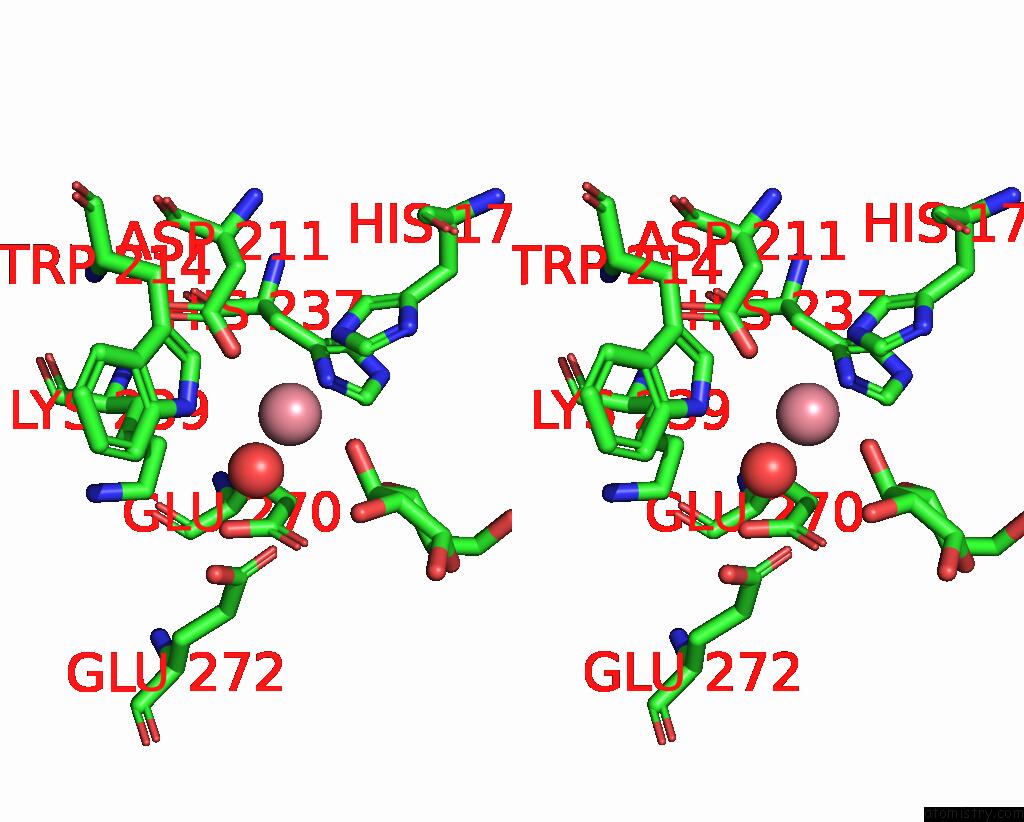
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 2 of Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1 within 5.0Å range:
|
Cobalt binding site 3 out of 4 in 8tcr
Go back to
Cobalt binding site 3 out
of 4 in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
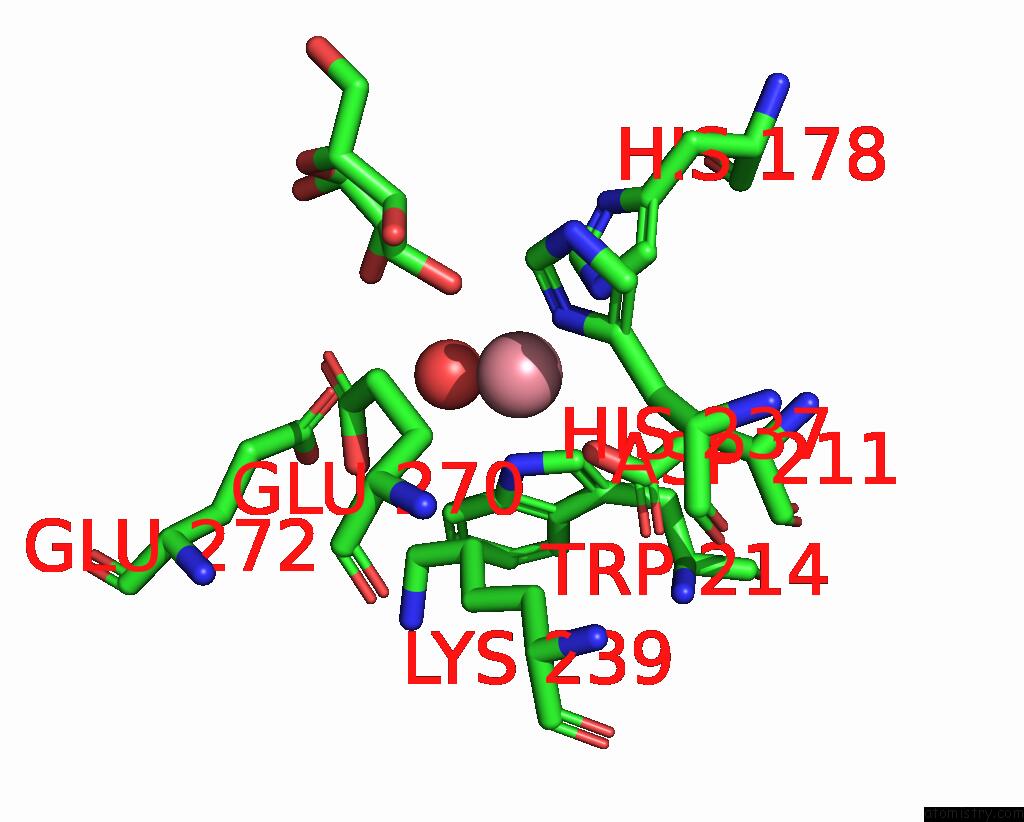
Mono view
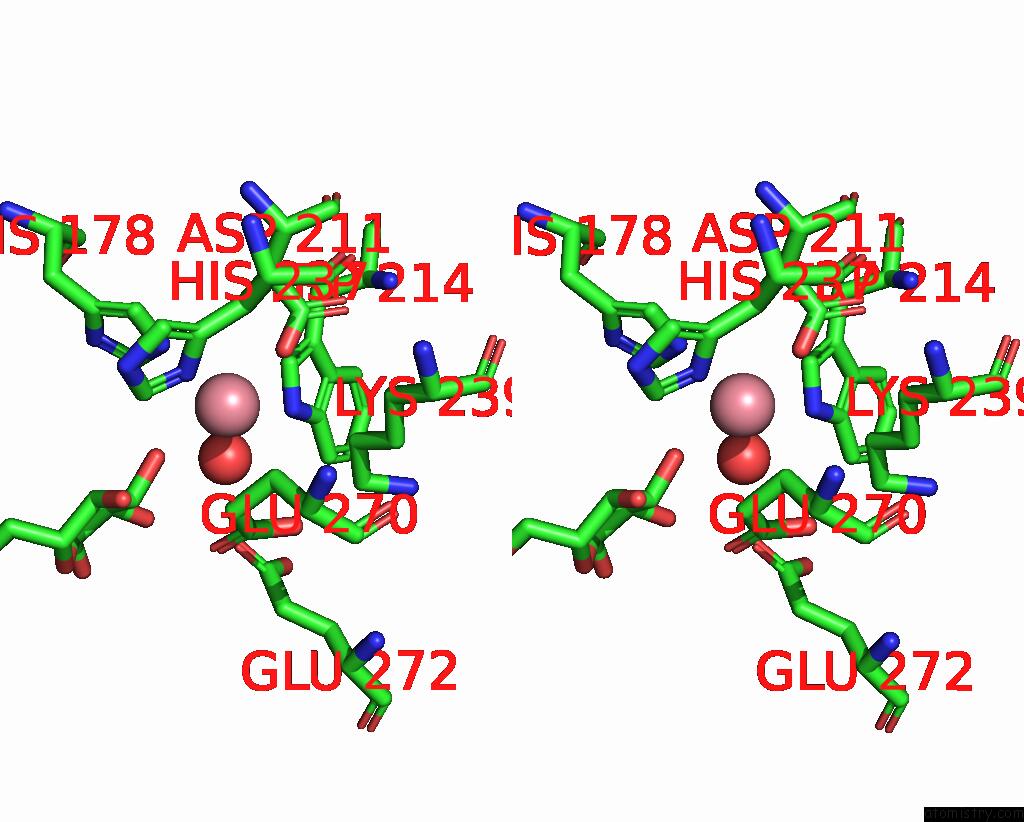
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 3 of Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1 within 5.0Å range:
|
Cobalt binding site 4 out of 4 in 8tcr
Go back to
Cobalt binding site 4 out
of 4 in the Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1
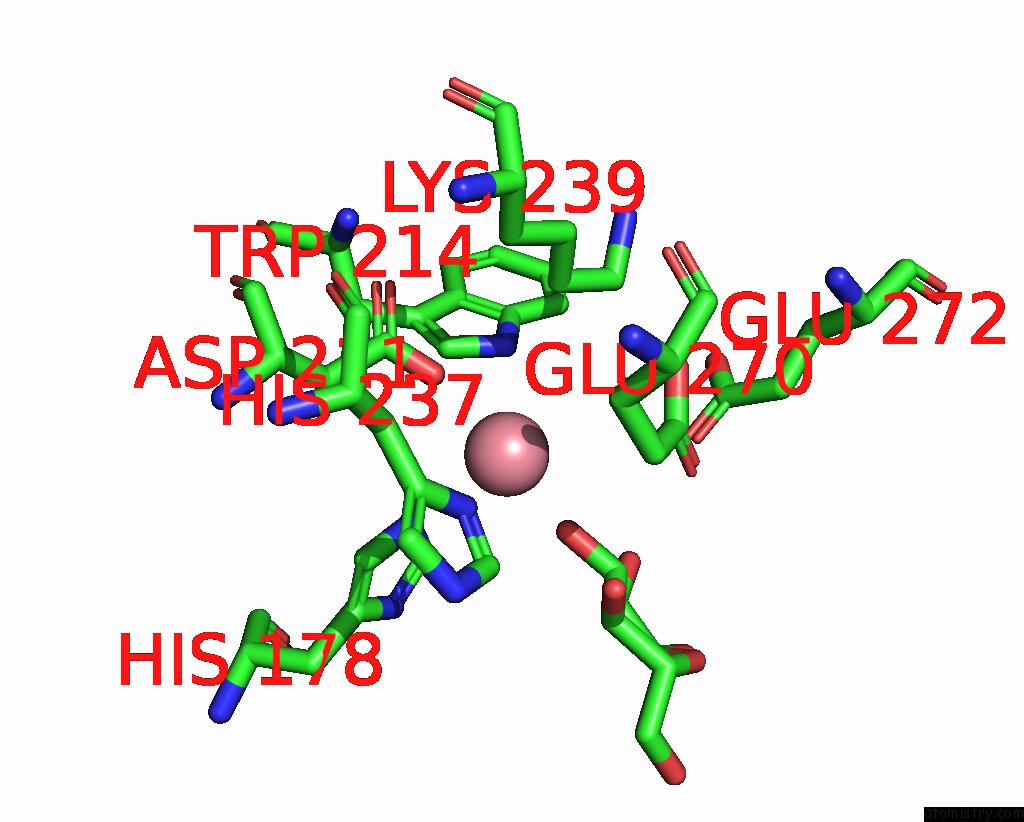
Mono view
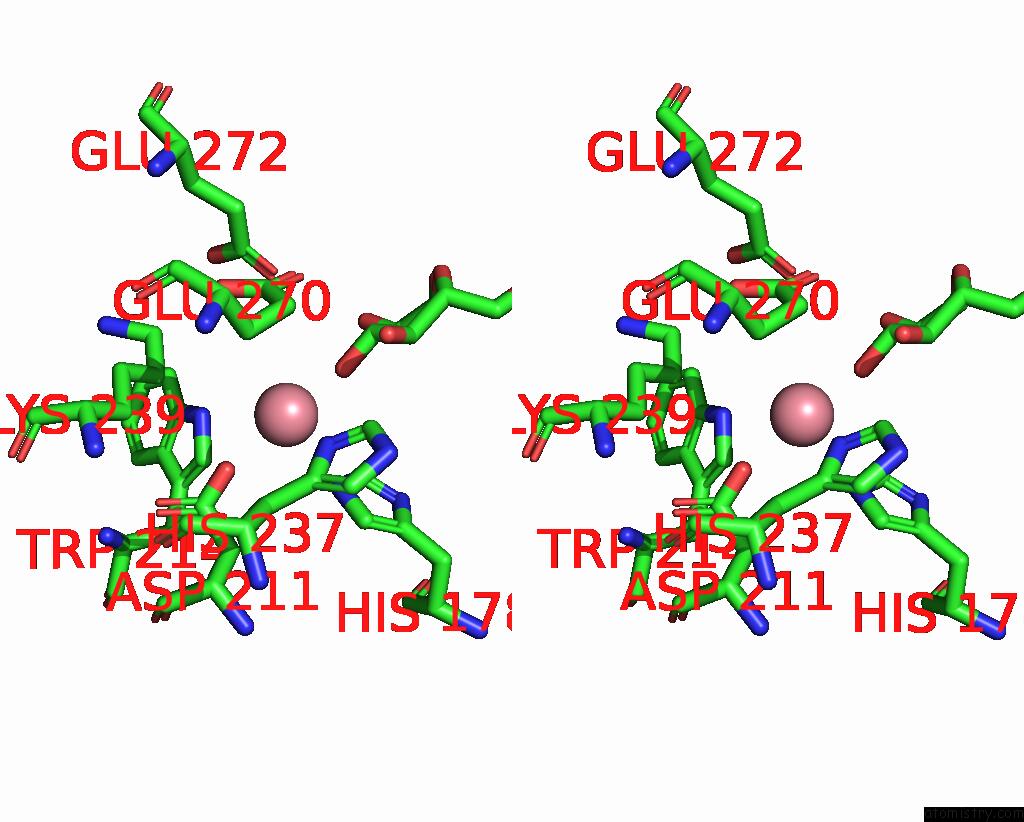
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 4 of Structure of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside- 1,2-Lyase AL1 within 5.0Å range:
|
Reference:
S.A.Nasseri,
A.C.Lazarski,
I.L.Lemmer,
C.Y.Zhang,
E.Brencher,
H.Chen,
L.Sim,
D.Panwar,
L.Betschart,
L.J.Worrall,
H.Brumer,
N.C.J.Strynadka,
S.G.Withers.
An Alternative Broad-Specificity Pathway For Glycan Breakdown in Bacteria Nature 2024.
ISSN: ESSN 1476-4687
Page generated: Sun Jul 13 22:06:00 2025
ISSN: ESSN 1476-4687
Last articles
Mg in 4DUXMg in 4DUW
Mg in 4DUV
Mg in 4DUO
Mg in 4DUG
Mg in 4DTY
Mg in 4DTW
Mg in 4DTH
Mg in 4DTF
Mg in 4DSU