Cobalt »
PDB 8xhj-9fvs »
9ehp »
Cobalt in PDB 9ehp: Co-Mahf-9 A8C Metal Alpha-Helix Framework
Protein crystallography data
The structure of Co-Mahf-9 A8C Metal Alpha-Helix Framework, PDB code: 9ehp
was solved by
R.M.Richardson-Matthews,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15.45 / 1.67 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 9.917, 16.292, 20.16, 81.61, 81.5, 72.51 |
| R / Rfree (%) | 20 / 24.9 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Co-Mahf-9 A8C Metal Alpha-Helix Framework
(pdb code 9ehp). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Co-Mahf-9 A8C Metal Alpha-Helix Framework, PDB code: 9ehp:
In total only one binding site of Cobalt was determined in the Co-Mahf-9 A8C Metal Alpha-Helix Framework, PDB code: 9ehp:
Cobalt binding site 1 out of 1 in 9ehp
Go back to
Cobalt binding site 1 out
of 1 in the Co-Mahf-9 A8C Metal Alpha-Helix Framework
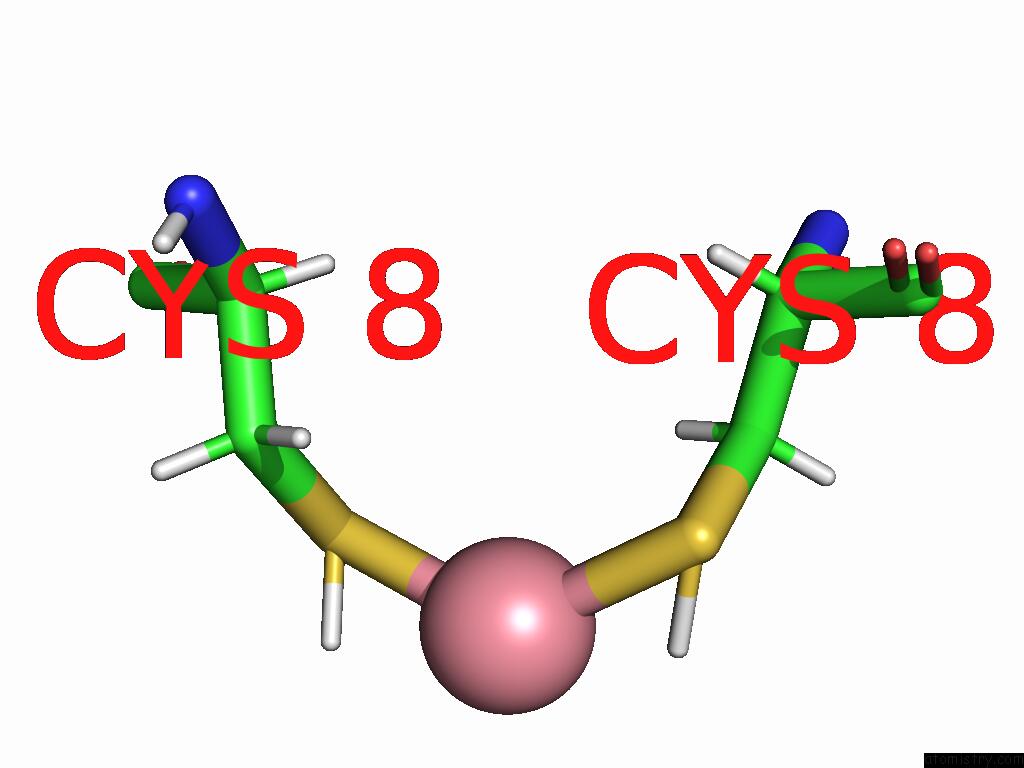
Mono view
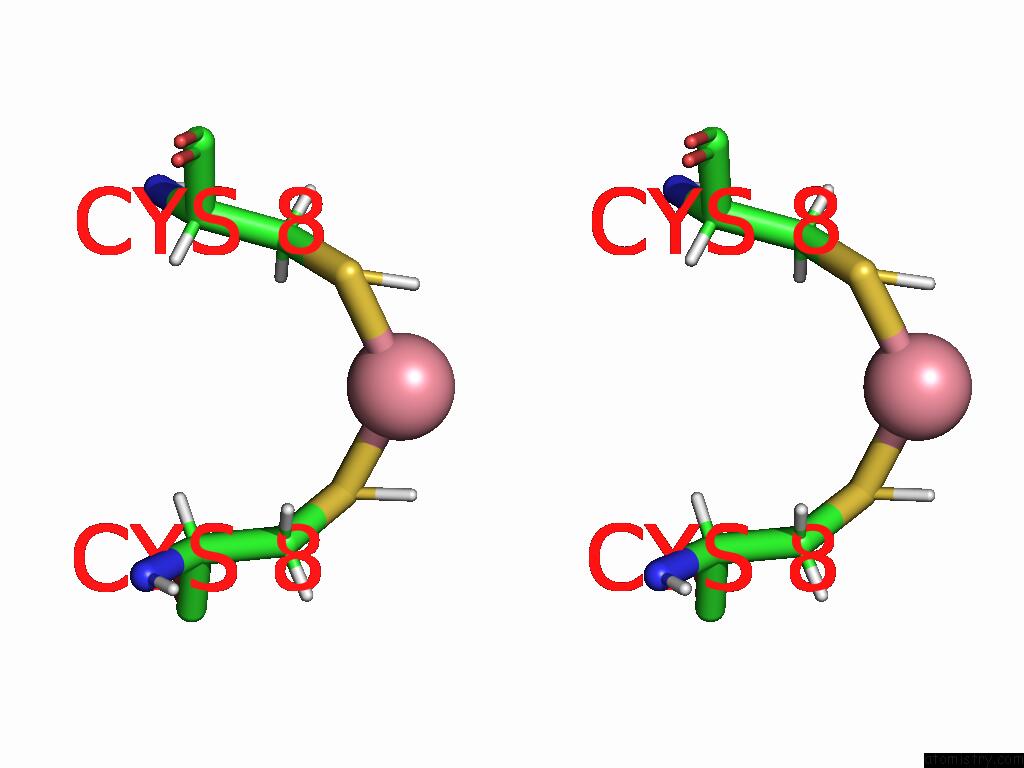
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Co-Mahf-9 A8C Metal Alpha-Helix Framework within 5.0Å range:
|
Reference:
R.Richardson-Matthews,
K.Velko,
B.Bhunia,
S.Ghosh,
J.Oktawiec,
J.S.Brunzelle,
V.T.Dang,
A.I.Nguyen.
Metal-Alpha-Helix Peptide Frameworks. J.Am.Chem.Soc. 2025.
ISSN: ESSN 1520-5126
PubMed: 40328673
DOI: 10.1021/JACS.5C04078
Page generated: Sun Jul 13 22:13:38 2025
ISSN: ESSN 1520-5126
PubMed: 40328673
DOI: 10.1021/JACS.5C04078
Last articles
Ir in 1ICGIn in 3VTM
In in 7WZ9
In in 1IND
I in 9NWF
I in 9KU6
I in 9M0I
I in 9RP9
I in 9KRR
I in 9IT8