Cobalt »
PDB 1e31-1hgw »
1f78 »
Cobalt in PDB 1f78: Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure
(pdb code 1f78). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure, PDB code: 1f78:
In total only one binding site of Cobalt was determined in the Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure, PDB code: 1f78:
Cobalt binding site 1 out of 1 in 1f78
Go back to
Cobalt binding site 1 out
of 1 in the Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure
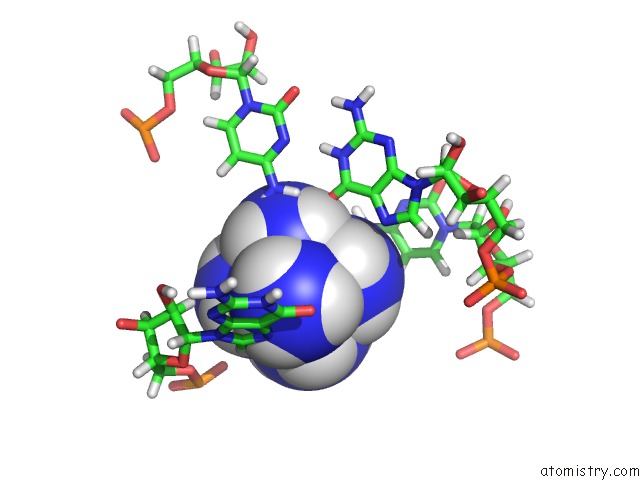
Mono view
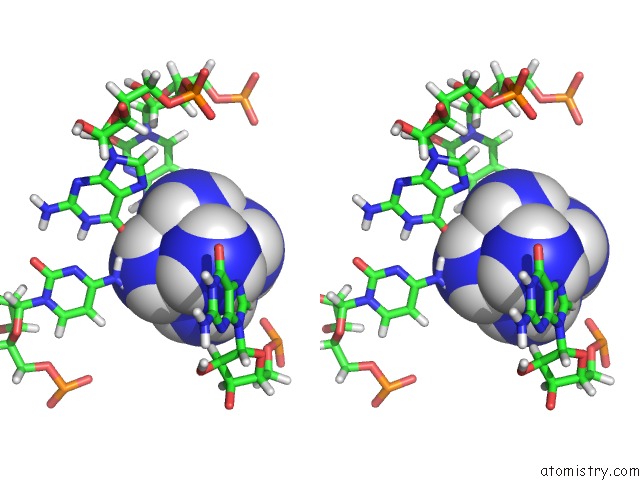
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Solution Structure of Rnase P Rna (M1 Rna) P4 Stem Oligoribonucleotide Complexed with Cobalt (III) Hexamine, uc(Nmr), Minimized Average Structure within 5.0Å range:
|
Reference:
M.Schmitz,
I.Tinoco Jr..
Solution Structure and Metal-Ion Binding of the P4 Element From Bacterial Rnase P Rna. Rna V. 6 1212 2000.
ISSN: ISSN 1355-8382
PubMed: 10999599
DOI: 10.1017/S1355838200000881
Page generated: Sun Jul 13 17:29:00 2025
ISSN: ISSN 1355-8382
PubMed: 10999599
DOI: 10.1017/S1355838200000881
Last articles
Cu in 4YSQCu in 4YSD
Cu in 4YSP
Cu in 4YSO
Cu in 4YSE
Cu in 4YSC
Cu in 4YSA
Cu in 4WTQ
Cu in 4YL4
Cu in 4XQZ