Cobalt »
PDB 1e31-1hgw »
1h0o »
Cobalt in PDB 1h0o: Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State
Enzymatic activity of Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State
All present enzymatic activity of Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State:
1.17.4.1;
1.17.4.1;
Protein crystallography data
The structure of Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State, PDB code: 1h0o
was solved by
K.R.Strand,
S.Karlsen,
K.K.Andersson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 8.0 / 2.2 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 75.900, 106.570, 91.620, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.5 / 28.2 |
Cobalt Binding Sites:
The binding sites of Cobalt atom in the Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State
(pdb code 1h0o). This binding sites where shown within
5.0 Angstroms radius around Cobalt atom.
In total only one binding site of Cobalt was determined in the Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State, PDB code: 1h0o:
In total only one binding site of Cobalt was determined in the Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State, PDB code: 1h0o:
Cobalt binding site 1 out of 1 in 1h0o
Go back to
Cobalt binding site 1 out
of 1 in the Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State
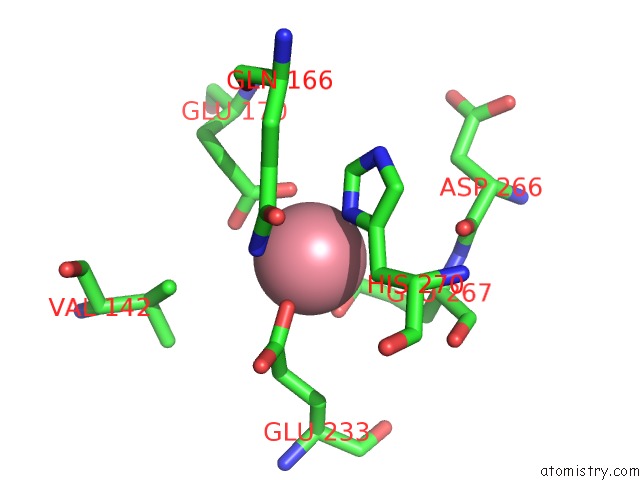
Mono view
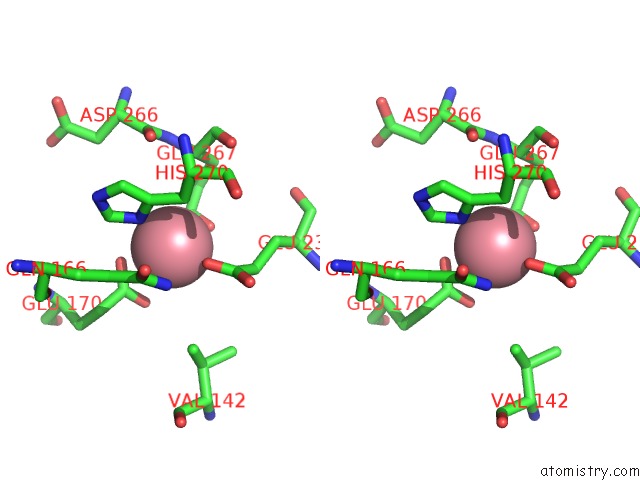
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cobalt with other atoms in the Co binding
site number 1 of Cobalt Substitution of Mouse R2 Ribonucleotide Reductase to Model the Reactive Diferrous State within 5.0Å range:
|
Reference:
K.R.Strand,
S.Karlsen,
K.K.Andersson.
Cobalt Substitution of Mouse R2 Ribonucleotide Reductase As A Model For Thereactive Diferrous State. Spectroscopic and Structural Evidence For A Ferromagnetically Coupled Dinuclear Cobalt Cluster J.Biol.Chem. V. 277 34229 2002.
ISSN: ISSN 0021-9258
PubMed: 12087093
DOI: 10.1074/JBC.M203358200
Page generated: Sun Jul 13 17:33:57 2025
ISSN: ISSN 0021-9258
PubMed: 12087093
DOI: 10.1074/JBC.M203358200
Last articles
Cu in 5AFACu in 5AKR
Cu in 5ACJ
Cu in 5ACI
Cu in 5ACH
Cu in 5ABD
Cu in 5ACG
Cu in 5ACF
Cu in 5A7E
Cu in 5AA7